1. IAMPDB Text Search
Facility
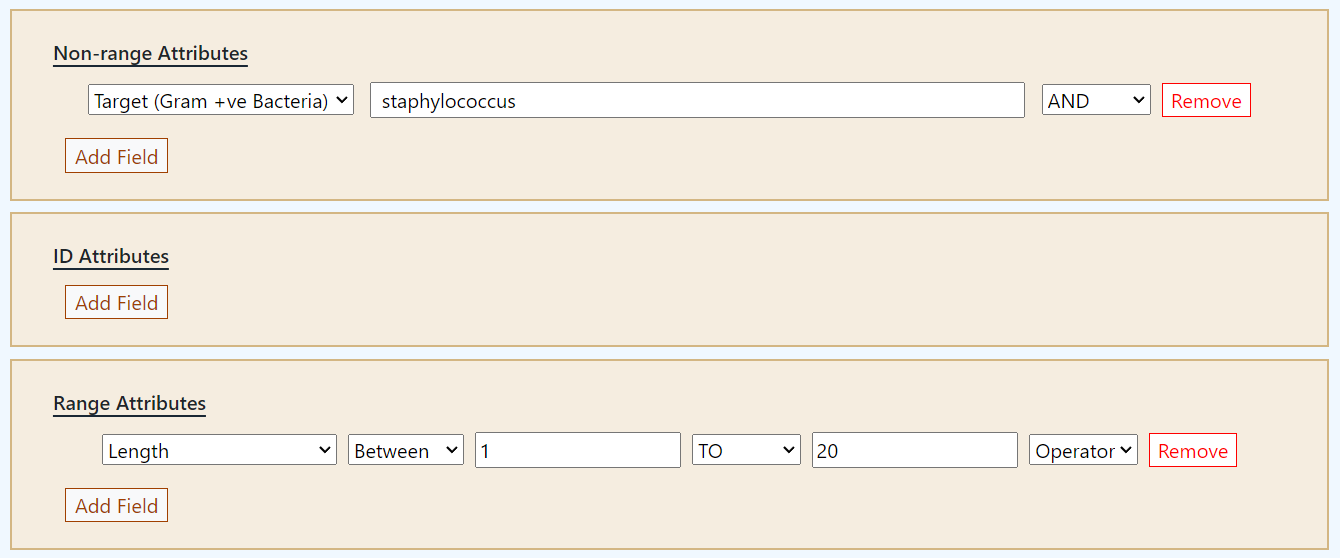
This is a generelized search facility.
This
search facility is case insensitive.
Here one can search with followings.
1. IAMPDB ID (e.g., IAMPDB10)
2. Peptide Name (e.g.,
Cecropin)
3. Source Organism [Insect only] (e.g., Drosophila melanogaster)
4. Taxonomy [Order] (e.g., Diptera)
5. Peptide Existence (e.g., Evidence at protein level)
6. Sequence (e.g., MNFYNIFVFVALILAITIGQSEAGWLKKIGKKIERVGQHTRDATIQGLGIAQQAANVAATARG) [Note: This is not a BLAST search]
7. Molecular Formula (e.g., C306H621N87O146S1)
8. Structural Class of the Peptide (e.g., alpha OR alpha_beta)
9. Antimicrobial Activities (e.g., Anti-gram-negative)
10. Enzymatic Activities (e.g., Hydrolase)
11. Inhibitory Activities (e.g., Enzyme inhibitor)
12. Other Biological Activities (e.g., Tumor suppressor)
13. Target Organism (Gram Positive Bacteria) (e.g., Staphylococcus aureus)
14. Target Organism (Gram Negative Bacteria) (e.g., Escherichia coli)
15. Target Organism (Fungi) (e.g., Candida)
16. Target Organism (Protozoa) (e.g., Plasmodium vivax)
17. Exception Target Organism (e.g., Serratia marcescens)
18. Binding Target (e.g., Cell membrane)
19. Related Assays Info (e.g., Inhibition Zone assay)
20. PubMed ID (e.g., 2104802)
21. UniProt ID (e.g., C0HKQ7)
22. PDB ID (e.g., 1D9O)